Research
Publications
Londschien M. and Bühlmann P. (2024) Weak-instrument-robust subvector inference in instrumental variables regression: A subvector Lagrange multiplier test and properties of subvector Anderson-Rubin confidence sets, arXiv:2407.15256
Londschien M., Bühlmann P. and Kovács S. (2023) Random Forests for Change Point Detection, Journal of Machine Learning Research
Londschien M., Kovács S., and Bühlmann P. (2021) Change point detection for graphical models in the presence of missing values, Journal of Computational and Graphical Statistics
Current projects
Distributional robustness of linear and tree-based models on ICU data: Train a model on a manually harmonized collection of ICU datasets to predict a clinical outcome such as length of stay. Which methods succeed in improving out-of-distribution performance when predicting on an unseen ICU dataset? What if we have a small sample of observations from the target dataset?
A foundation model for ICU data: Train a deep model on a manually harmonized collection of ICU datasets to predict future measurements. Can the resulting representation be used to predict clinical outcomes? How do predictions generalize to unseen ICU datasets?
Drift and batch correction of metabolomics data: Metabolites are small molecules. Body fluid samples (e.g., blood) of different patients are aggregated on plates and metabolite counts are measured sequentially by mass spectrometry. These measurements incur drifts and batch effects. Can we use splines to remove the drifts? Finding batch boundaries is a change point localization problem. Are the metabolite measurements after correction with our method more predictive for some phenotypes than the uncorrected measurements?
Weak-instrument-robust subvector inference in instrumental variables regression: A subvector Lagrange multiplier test and properties of subvector Anderson-Rubin confidence sets
Londschien M. and Bühlmann P. (2024), arXiv:2407.15256
Instrumental variables regression (IV) is a method for causal effect estimation when controlled experiments are infeasible. If the explanatory variable of interest is correlated with the error term (endogenous), standard regression techniques such as ordinary least squares (OLS) yield biased estimates for the causal effect. IV uses instruments, variables that are correlated with the explanatory variable but not with the error, to isolate variation in the explanatory variable of interest that is uncorrelated with the error term, to yield an unbiased estimate of the causal effect.
To influence policy, uncertainty quantification is essential. Typically, Wald tests based on the estimator’s asymptotic normality are used to construct confidence sets and p-values. In many applications of IV, the signal-to-noise ratio in the first stage, from the instruments to the explanatory (endogenous) variable of interest, is low, leading Wald-based confidence sets to undercover.
An alternative are tests that are robust to such weak instruments.
The Anderson-Rubin test is one option. It has the correct size independently of instrument strength and confidence sets obtained by test inversion have a closed-form solution. However, the degrees of freedom of the limiting chi-squared distribution are equal to the number of instruments, not the number of tested parameters. Consequently, adding instruments might lead to larger confidence sets.
Another weak-instrument-robust test is Kleibergen’s (2002) Lagrange multiplier test. The degrees of freedom of its limiting chi-squared distribution equal the number of parameters under test. Consequently, adding instruments should only improve identification and result in smaller confidence sets.
If there are multiple endogenous variables, confidence sets for the entire parameter vector are difficult to interpret. Subvector confidence sets for individual coefficients of the causal parameter are more useful. Guggenberger et al. (2012) propose a subvector variant of the Anderson-Rubin test by plugging in the LIML (the MLE assuming Gaussian noise) for the nuisance parameter. In the same paper they also show that the similarly obtained subvector variant of the Lagrange multiplier test has incorrect size.
In our work, we
- propose a subvector variant of the Lagrange multiplier test by minimizing over the nuisance parameter. We show that this has the correct size under a technical condition. This is the first weak-instrument-robust subvector test in instrumental variables regression to recover the degrees of freedom of the (not weak-instrument-robust) Wald test.
- We compute the closed-form solution for the confidence sets obtained by inverting the (subvector) Anderson-Rubin test. We show that the $1 - \alpha$ subvector confidence sets for an individual parameter are jointly bounded if and only if Anderson’s (1951) likelihood ratio test that the first-stage parameter is of reduced rank rejects at level $\alpha$. The confidence sets are centered around a k-class estimator and, if bounded and non-empty, equal a Wald-based confidence set around that estimator.
Our test fills a gap in the weak-instrument-robust testing literature for instrumental variables regression:
| Anderson-Rubin | Lagrange multiplier | conditional likelihood-ratio | |
|---|---|---|---|
| complete | Anderson and Rubin (1949) | Kleibergen (2002) | Moreira (2003) |
| subvector | Guggenberger et al. (2012) | us | Kleibergen (2021) |
Let $k = \mathrm{dim}(Z)$ be the number of instruments and $m \leq k$ be the number of endogenous variables. Subvector tests are for a single parameter.
| degrees of freedom | subvector DOF | weak-instrument-robust? | closed-form confidence set? | possibly unbounded? | |
|---|---|---|---|---|---|
| Wald | $m$ | $1$ | no | yes | no |
| likelihood-ratio | $m$ | $1$ | no | yes | yes |
| Anderson-Rubin | $k$ | $k - m + 1$ | yes | yes | yes |
| conditional likelihood-ratio | in-between | in-between | yes | no | yes |
| Lagrange multiplier | $m$ | $1$ | yes | no | yes |
I implemented all the (subvector) tests mentioned above and the (subvector) confidence sets obtained by test inversion in the open-source python sofware package ivmodels. See also the docs. This also implements k-class estimators, such as the two-stage least-squares (TSLS), limited information maximum likelihood (LIML), and Fuller estimators, and auxiliary tests such as the J-statistic of overidentification and Anderson’s (1951) likelihood-ratio test of reduced rank. A summary containing estimates test statistics, p-values, and confidence sets can be generated with the k-class estimator’s summary method. ivmodels is available on conda-forge and pypi.
See also the examples in the docs:
- Card (1995): Using Geographic Variation in College Proximity to Estimate the Return to Schooling
- Tanaka, Camerer, and Nguyen (2010): Risk and Time Preferences: Linking Experimental and Household Survey Data from Vietnam
- Angrist and Krueger (1993): Does Compulsory School Attendance Affect Schooling and Earnings?
- Acemoglu, Johnson, Robinson (2001): The Colonial Origins of Comparative Development: An Empirical Investigation
The paper also includes a selective review of the literature on weak-instrument-robust inference for instrumental variables regression in the appendix.
Random Forests for Change Point Detection
Londschien M., Bühlmann P. and Kovács S. (2023), Journal of Machine Learning Research
Change point detection considers the localization of abrupt distributional changes in ordered observations, often time series. Parametric methods typically assume that the observations between the change points stem from a finite-dimensional family of distributions. Change points can then be estimated by maximizing a regularized log-likelihood over various segmentations.
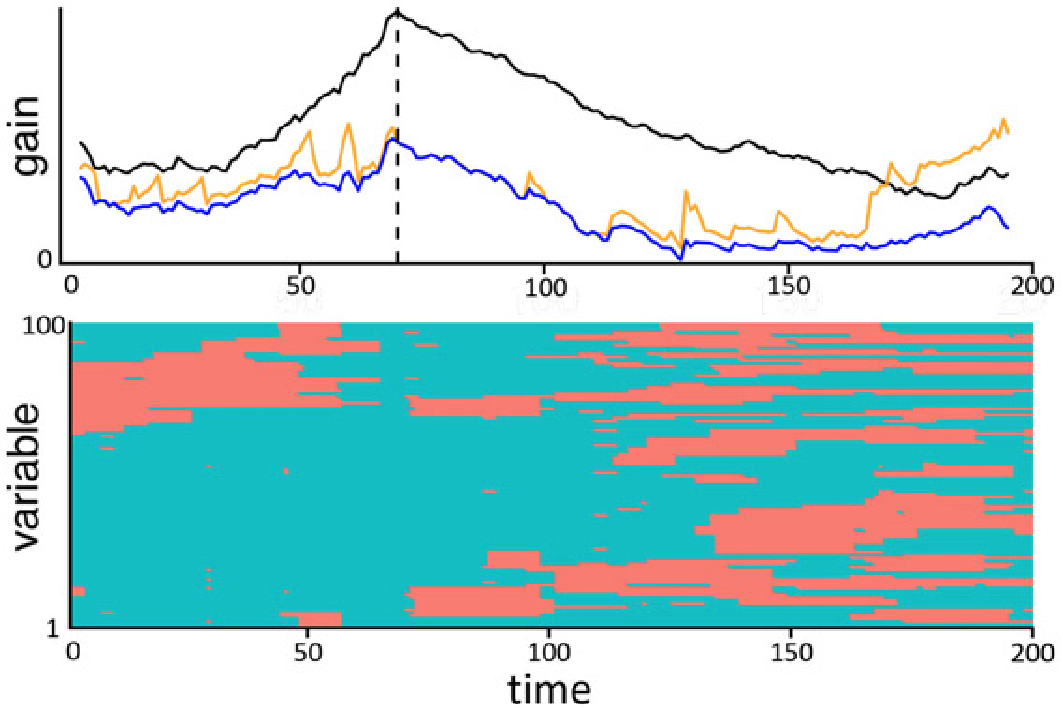
See below for an illustration with Gaussian data and a single change in mean. The data is $x_i \sim \mathcal{N}(-1, 1)$ for $i \leq 80$ and $x_i \sim \mathcal{N}(2, 1)$ for $i > 80$. For each split $s = 20, \ldots, 180$, we fit two Gaussians on $x_1, \ldots, x_s$ and $x_{s+1}, \ldots, x_{200}$ and compute the resulting log-likelihood. On the right is, for each split point $s$, the improvement in log-likelihood (gain) when splitting at $s$, compared to a single Gaussian fit for all data points $x_1, \ldots, x_{200}$. This is often called the CUSUM.
Nonparametric change point detection methods use measures that do not rely on parametric forms of the distribution or the nature of change. We propose to use classifiers to construct a novel class of multivariate nonparametric multiple change point detection methods.
Intuitively, if there is a change point at time point $t_{\mathrm{change}}$, a classifier should perform better than random at separating observations from before $t_{\mathrm{change}}$ from observations after $t_{\mathrm{change}}$. We propose a classifier likelihood-ratio that uses class probability predictions to compare different change point configurations. We combine this with a search method based on binary segmentation and prove that this consistently recovers a change point when covered with a classifier that provides consistent probability estimates.
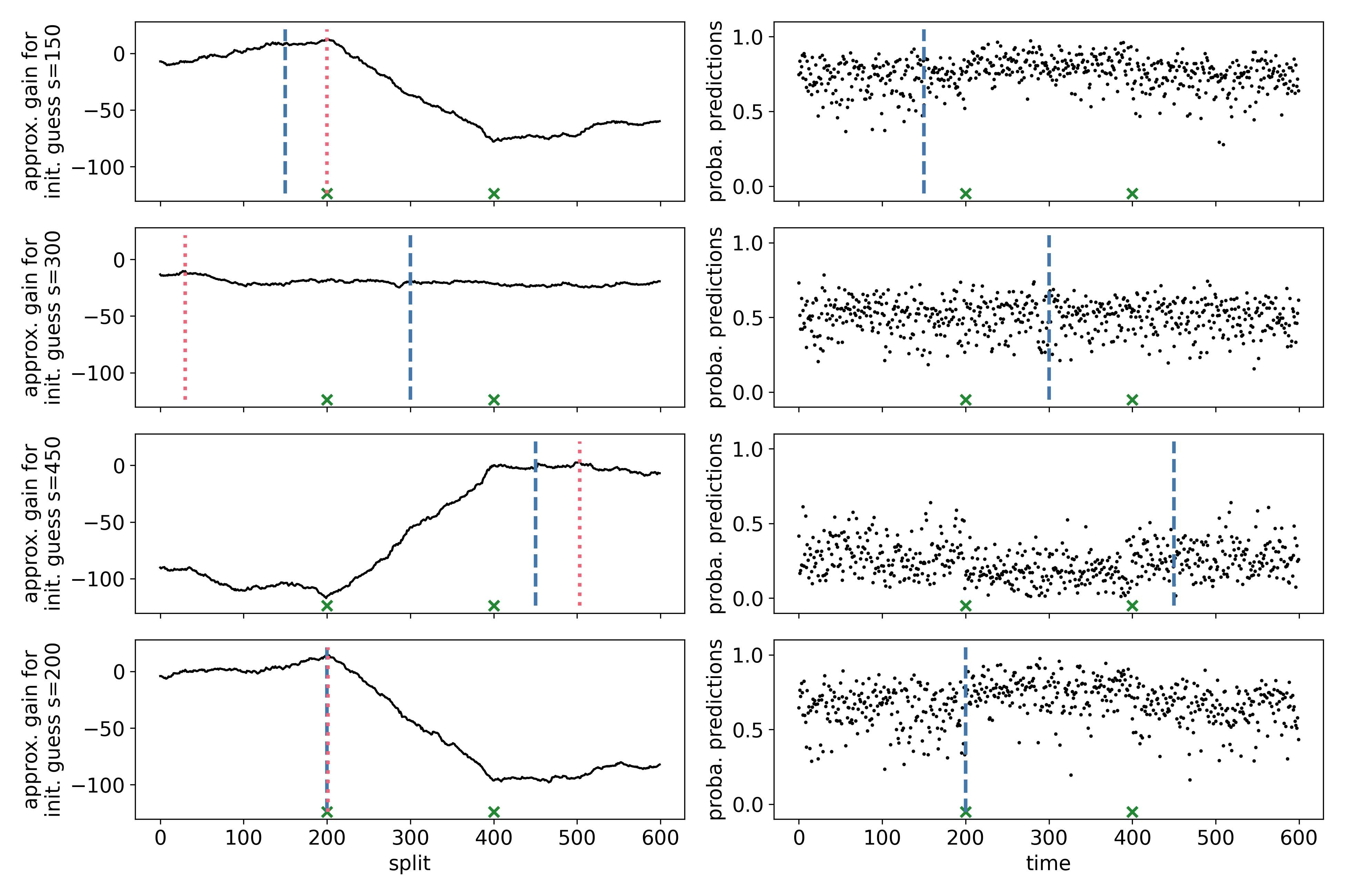
The figure below is based on a synthetic dataset with change points at $t = 200, 400$. The distributions of the first and last segment are equal. We assign a label 0 to observations left to the “first guess” (blue dashed line) and a label 1 to the right. The right panels contain the out-of-bag probability predictions of a random forest. These probability predictions yield classifier log-likelihood ratios, which aggregate to a “gain” in the left panel, similar to the CUSUM above. The first 3 panels contain gain curves for different “first guesses”. The last panel contains a gain curve using the maximizer of the first 3 gain curves as an initial guess.
The method is implemented in the changeforest software package available in R (conda-forge), Python (conda-forge and pypi), and Rust (crates.io).
Below is a video of a bee doing the “waggle dance” to communicate the direction and distance to a collection of pollen.
We fit changeforest on the differences of the green box’s x, y-coordinates, and angle for each frame.
In : changeforest(dX)
Out: best_split max_gain p_value
(0, 180] 41 9.793 0.005
¦--(0, 41] 19 -3.148 0.36
°--(41, 180] 106 9.385 0.005
¦--(41, 106] 82 -3.831 0.16
°--(106, 180] 153 21.758 0.005
¦--(106, 153] 124 0.314 0.08
°--(153, 180]
The changeforest algorithm detects changepoints at $t=41, 106, 153$, corresponding to the time points where the bee changes between “waggling” and rotating.
Change point detection for graphical models in the presence of missing values
Londschien M., Kovács S. and Bühlmann P. (2021) Journal of Computational and Graphical Statistics
Most research in change point detection assumes the availability of complete observations. However, in many applications, some of the observations might be missing, with a missingness pattern that is not completely at random. This leads to a kind of “chicken and egg problem”: Missing values need to be imputed before standard change point methods can be applied. But, most imputation methods assume i.i.d. data, so to apply them, knowledge of the change points is required. When one imputes data as if the data was i.i.d., and if the missingness pattern is not missing completely at random, a subsequently applied change point algorithm possibly detects changes in the missingness pattern, not the underlying data.
We propose an approach that integrates the missingness directly into the change point detection loss function, assuming data is generated from a sparse Gaussian graphical model. We fit a covariance structure using the graphical Lasso. If, for a given segment, one variable has too many missing entries, we do not consider that variable.
For the following data we simulated data with “blockwise missing” data, visualized in red on the bottom. The black line is the gain if the full data without missing values is used. The gain of the naive approach with simple imputation over the entire data is in orange. Our approach is in blue, with a maximum close to the true underlying change point (blue dashed vertical line).